Getting started¶
Clone a local copy of this repository from Github using:
git clone https://github.com/MarcusJP/ODYNN
Go to the root directory and run:
make init
make install
Some examples¶
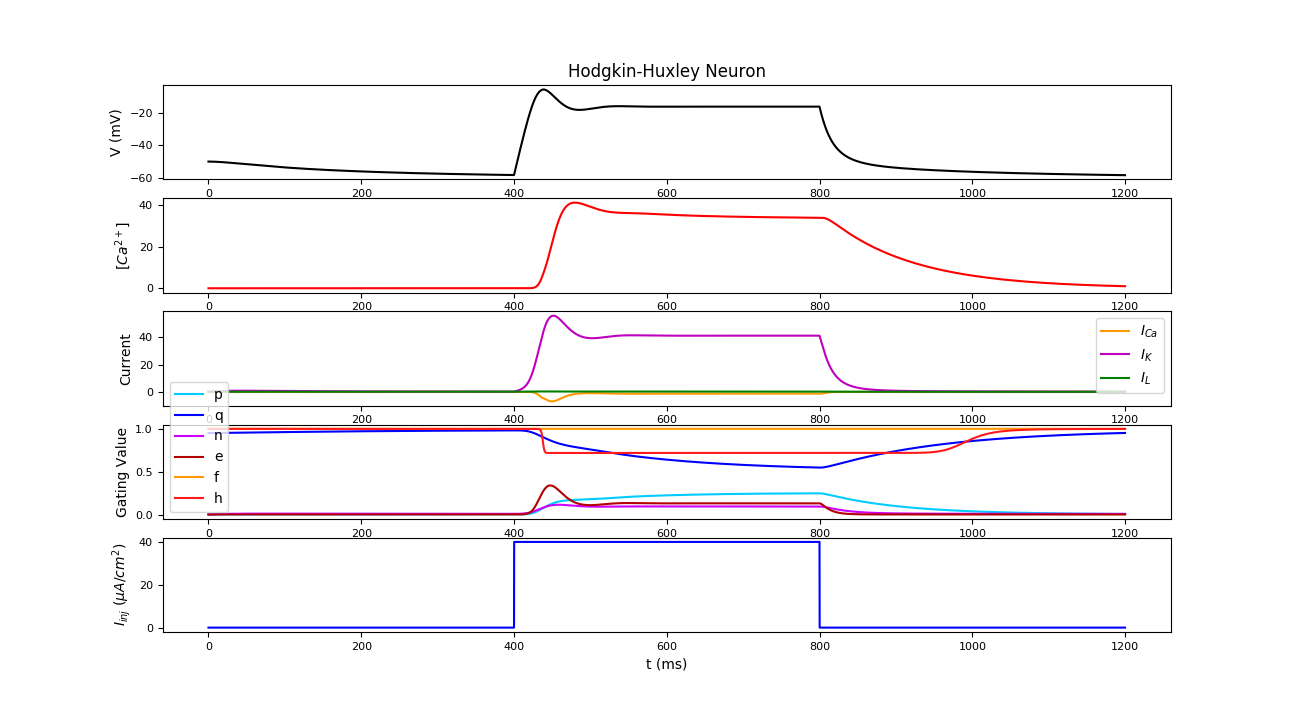
- Neuron simulation :
from odynn.nsimul import simul
import scipy as sp
t = sp.arange(0., 1200., 0.1)
i = 20. * ((t>400) & (t<800))
simul(t=t, i_inj=i, show=True)
- Neuron optimization :
import numpy as np
from odynn import utils, nsimul, neuron, noptim
#This file defines the model we will use
from odynn.models import cfg_model
dt = 1.
folder = 'Example'
# Function to call to set the target directories for plots and saved files
dir = utils.set_dir(folder)
#Definition of time and 2 input currents
t = np.arange(0., 1200., dt)
i_inj1 = 10. * ((t>200) & (t<600)) + 20. * ((t>800) & (t<1000))
i_inj2 = 5. * ((t>200) & (t<300)) + 30. * ((t>500) & (t<1000))
i_injs = np.stack([i_inj1, i_inj2], axis=-1)
#10 random initial parameters
params = [cfg_model.NEURON_MODEL.get_random() for _ in range(10)]
neuron = neuron.BioNeuronTf(params, dt=dt)
#This function will take the default parameters of the used model if none is given
train = nsimul.simul(t=t, i_inj=i_injs, show=True)
#Optimization
optimizer = noptim.NeuronOpt(neuron)
optimizer.optimize(dir=dir, train=train)
Tutorials¶
Jupyter notebook tutorials are available. After cloning the repository, go to the tutorial/ folder and run:
jupyter notebook